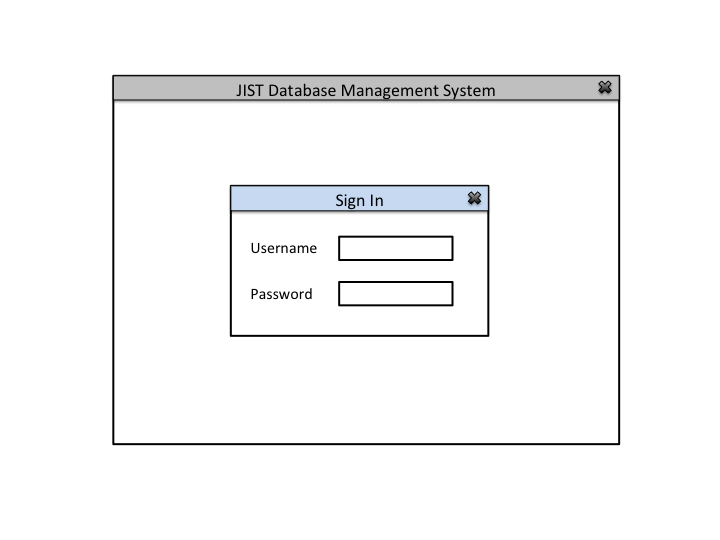
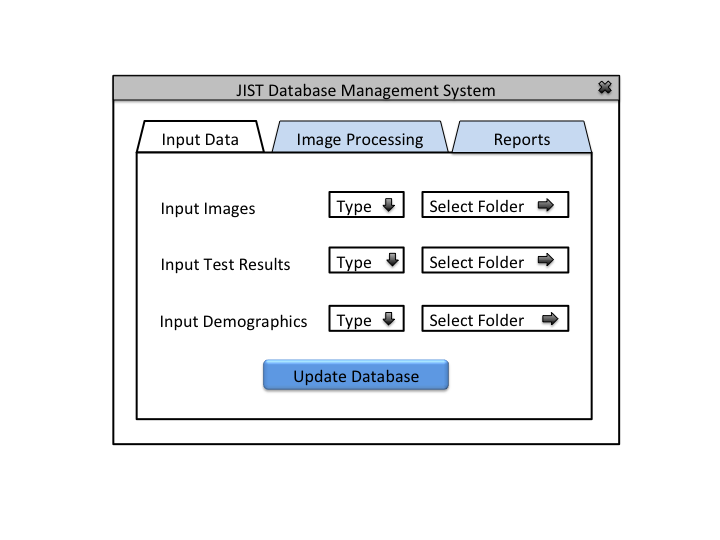
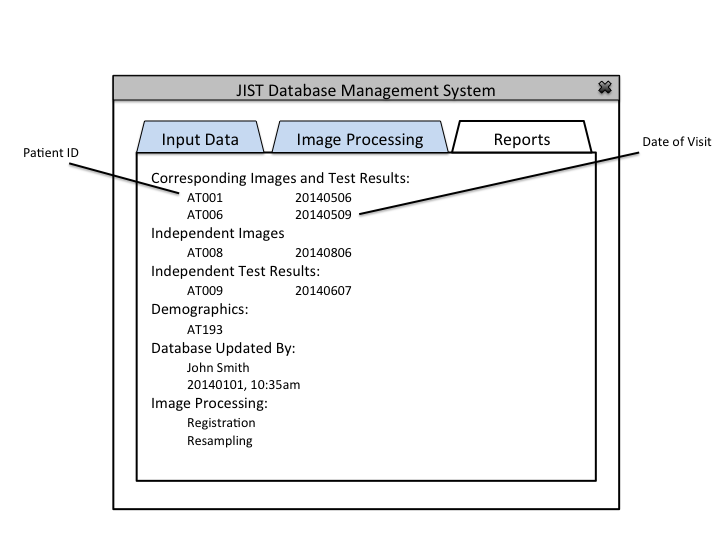
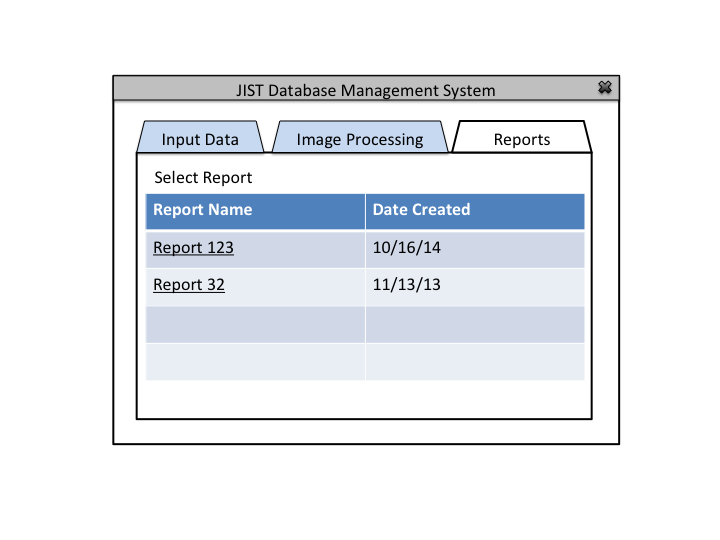
This application will allow the users (IACL, JHU) to collect, organize, and use medical image and experiment data for research purposes. It is challenging to organize and maintain medical data because of the longitudinal nature of the studies, inconsistent data collection procedures, and the constant influx of new datasets.
Normally, when a patient schedules a visit, they go through medical examinations followed by a medical imaging procedure performed by different groups of people (imaging and testing can be done individually or within short intervals as well). Researchers are often interested in analyzing the image data with a corresponding examination result. Therefore it is important to organize the data in a way that makes it easier to associate related data (related by visit, patient, family members, diagnosis, etc.).
We are aiming to:
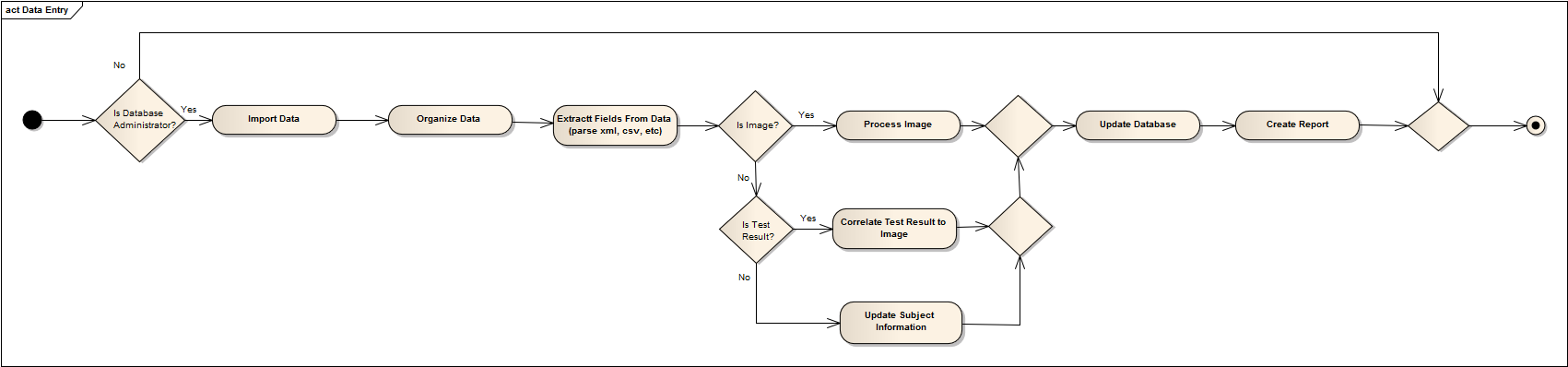
1- Develop a database management system that could parse, restructure, and organize different types of medical data through an intuitive user interface. We want to incorporate some basic image processing with the database system to create more applicable data from the incoming datasets. Please note that database management can only be done by authorized personnel.
2- Develop an interface to retrieve desired data from the dataset. This can be incorporated into the JIST (Java Image Science Toolkit) framework as a module.
3- Develop a database version control system (strech goal)
Primary Actor: Database Administrator
Secondary Actor: Database
Goal: Insert data into the database
Primary Actor: Database
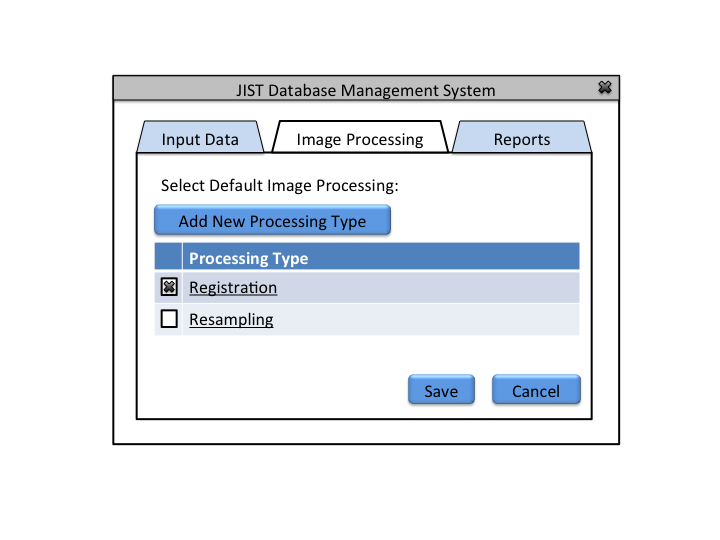
Goal: Apply basic image processing and the user specified default image processing to images being inserted into the database
Primary Actor: Database Administrator
Secondary Actor: Database
Goal: Define the default image processing
Primary Actor: User
Secondary Actor: Database
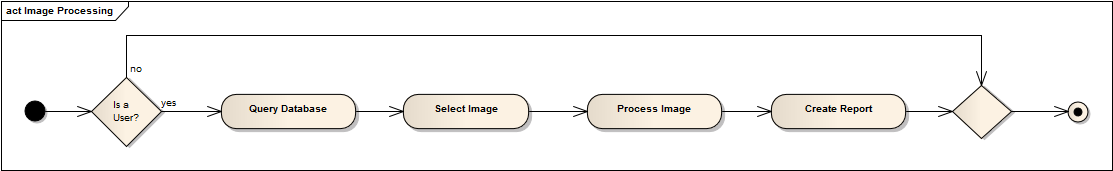
Goal: Query database for necessary data processing
The Database Administrator is used to maintain and update the database. Authorization is required. Generates a report after each update.



The Database Administrator can also view past reports and define default image processing
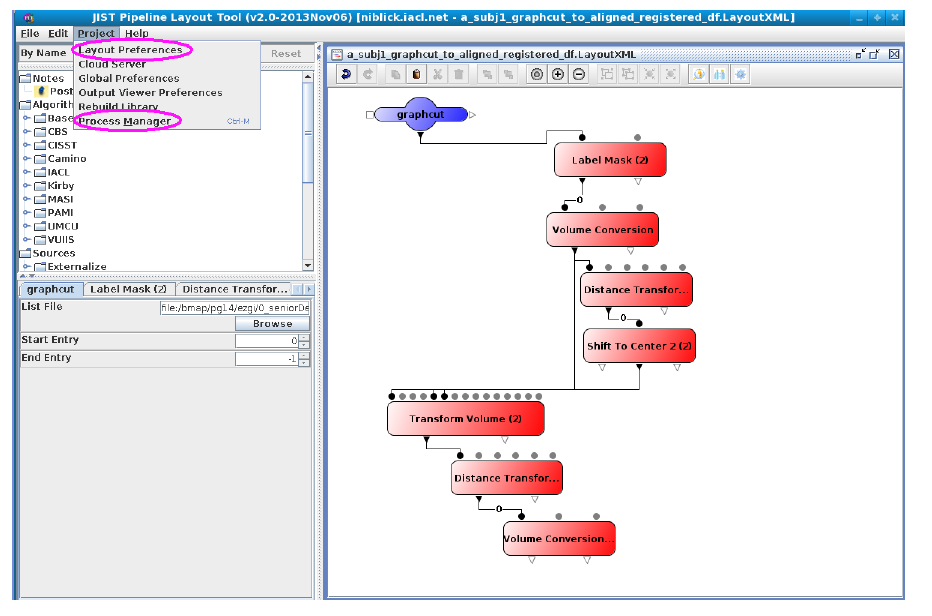
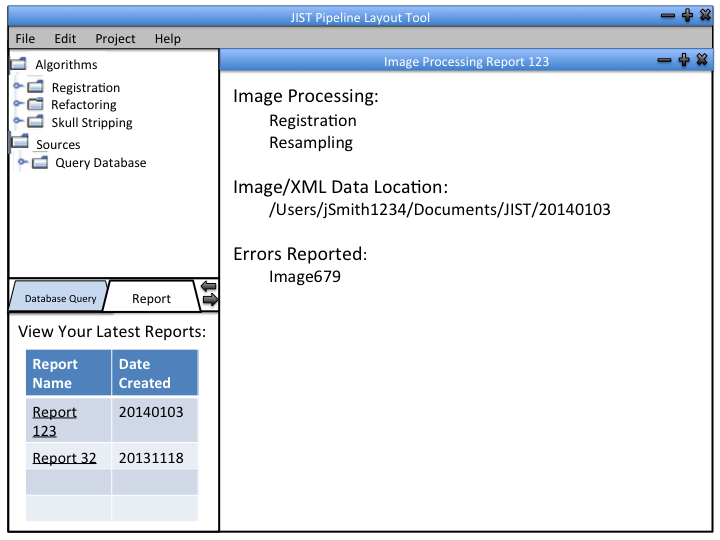
The image processing pipeline is implemented using Java Image Science Toolkit (JIST). JIST is a Java-based imaging processing environment which was developed as an extension to MIPAV (Medical Image Processing, Analysis, and Visualization). Using this environment, multiple modules that contain one or more algorithms can be connected into a pipeline to process the data. Output from one module is connected to another to create a continuous flow of analysis. The pipelines can be distributed using layout files (.LayoutXML). The user interface looks like this:
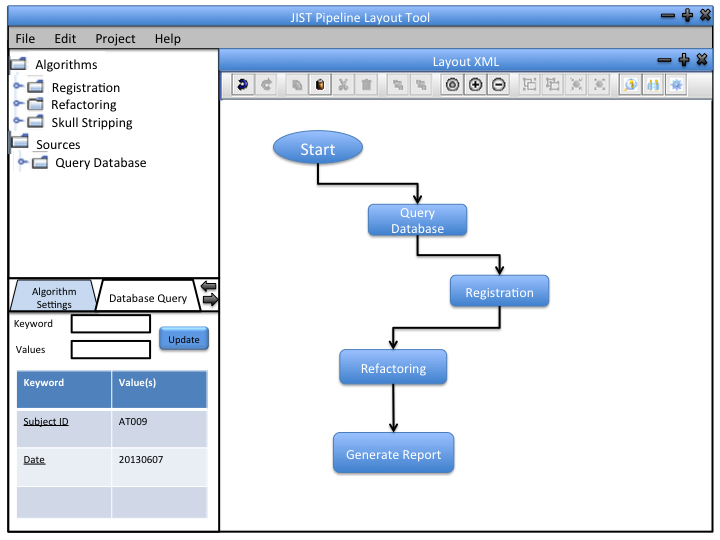
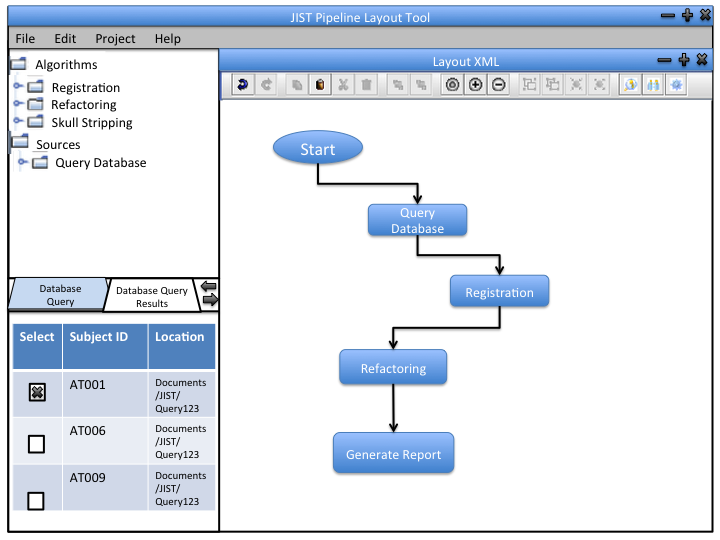
We can incorporate out data retrieval into the JIST framework so that the user can integrate the software into their pipeline designs. The module can be used independently to produce output files (.txt) that contain desired information from the database.
Please click here for the JavaDoc Documentation
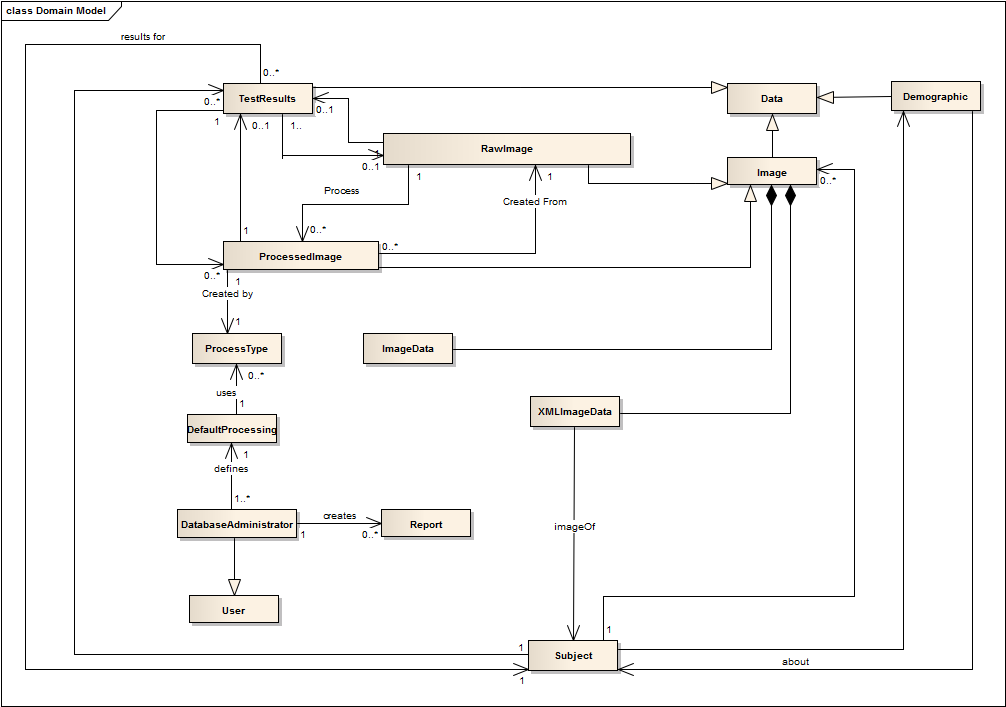
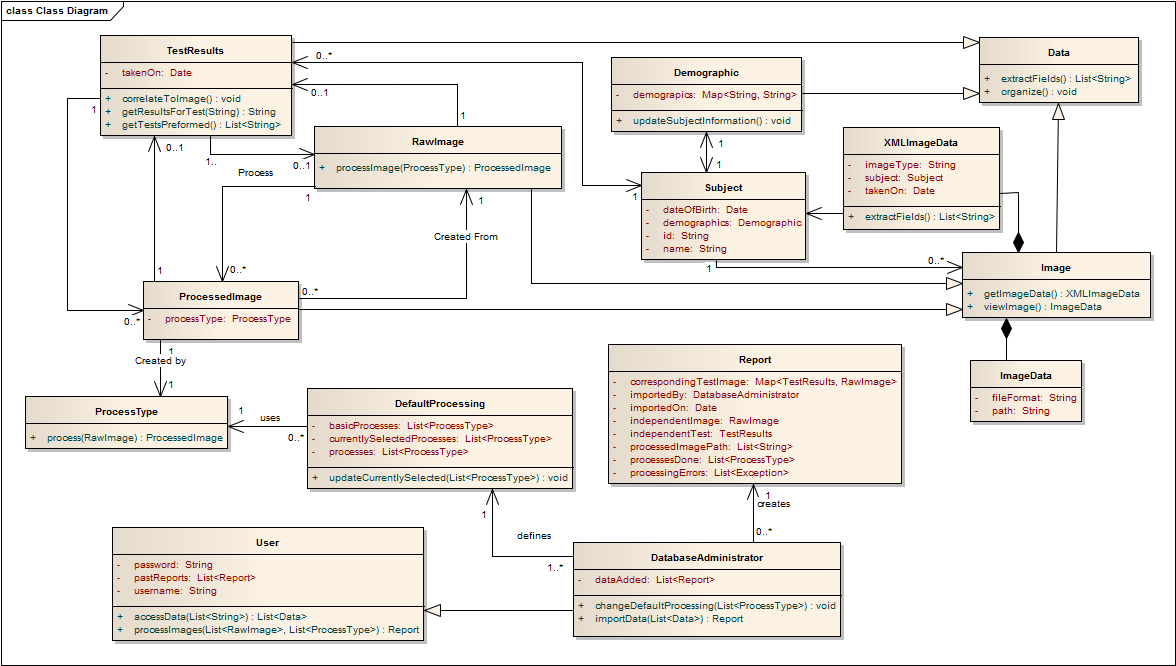
We didn't include component diagram here, because in the concept of our software the number of nodes are limited, and the relationships are obvious from class diagram.
A basic client server interaction is used for this project. The client-side application allows a user to connect via own computer to the database server. The server is running Hibernate, MySQL, and JIST functions for image processing and storage. The results for processing jobs are stored on the server-side. The two main packages that we will implement includes one for the client and one for the server.
We are going to build our project on an existing Java based image processing and analysis environment (JIST). JIST is a framework for algorithm development and large-scale image processing which combines a fully functional, graphical pipeline environment with a framework to take advantage of advanced multi-modality imaging libraries available within Java. JIST integrates closely with MIPAV (Medical Image Processing, Analysis and Visualization, National Institutes of Health), a widely used and well-supported multidimensional imaging, visualization, and processing package from the National Institutes of Health which provides for user-friendly visualization and exploration of the multi-dimensional imaging data as well as three-dimensional structures.
JIST has a layout panel which provides a graphical interface to arrange image processing algorithms as modules (see GUI sketches). Modules can be dragged and dropped from the module panel into the layout panel. When a module is selected in the layout panel, a dialog appears in the parameter panel that allows the user to edit input parameters for that module [Lucas, Blake C, et al. The Java Image Science Toolkit (JIST) for rapid prototyping and publishing of neuroimaging software, Neuroinformatics, 2010 Mar].
Using this environment, multiple modules that contain one or more algorithms can be connected into a pipeline to process the data. Output from one module is connected to another to create a continuous flow of analysis. The pipelines can be distributed using layout files (.LayoutXML). There are 2 parts to our project:
1 - Database management: This may or may not be incorporated into JIST framework. It will use Hybernate to update the database.
2 - Interaction with the database: This will be implemented as a JIST module (like an image processing module). This way, users can incorporate it into their image processing pipelines. Users can specify the data they want from the database using the parameter panel.