Object Oriented Software Engineering Course - Group 8
Jeff Burka (Frontend)
Bumjin Kim (Backend, Frontend)
Alex Monroe (Frontend)
Emily Wagglestone (Backend)
Title : OliGUI
Architecture
The project will use ruby programming language and ruby on rails 4.1.6 which is built on top of ruby 2.1. The backend will be handled by the ruby on rails framework while the frontend contains html, javascript langauges. The frontend frameworks include jQuery. We will use hosting service to deploy the website. The ruby on rails includes major components. One of which is Action Pack which contains action controller, action dispatch, and action read.
Resources
We will be using ruby on rails framework as a foundation for this project. The packages that might be used are Easy_downloader 0.1.1 and salted_login_generator packages. The salted_login_generator package generates the login system of the ruby on rails website. We will use Ruby on Rails API for ActionModel, ActionView and ActionController. For action controller, ActionController:Cookies to store information in the cookie. For action view, ActionView::Renderer to render the web.
Vision Statement
Our project will be a web app that will allow users to do drag-and-drop DNA design. We will pull from an existing bioparts database as the supply for BioBricks (DNA building blocks with specific functions). Users will be able to drag and drop BioBricks together to create a large DNA sequence. The supply will be indexed by BioBrick ID number, which the user will be able to use in order to find the pieces they are looking for. In addition, BioBricks are sorted into different categories (promoter, encoder, terminator, etc.) in order to further facilitate search. Once the user finishes building the sequence, a simple parser will run to make sure the strand makes sense. For example, the parser will notify the user when the number of promoter regions does not match the number of terminator sequences. After the simple parsing, our program will be able to perform simple codon optimization based on the organism that the strand is targeted for. We will either use a simple optimization algorithm that we build from scratch or push the sequence to an external codon optimizer. Once the sequence is fully constructed, our website will allow users to save and download the final sequence, save their sequence to a library associated with their account, and send their sequence out to oligo manufacturing companies for processing. In order to save the sequence to an account, we will need to allow users to create an account with the website that will be kept on the server. The backend of the website will use Rails, while the front end user interface will be written in Javascript.
Developed out of MIT, BioBricks are essentially DNA Legos. While its often difficult to determine the function of a stretch of DNA, BioBricks are highly-studied sequences. Because we are certain of what their exact functions, we can plug them together to build more complex sequences to do more interesting things within a cell. There are BioBricks for everything from producing Vitamin C to cellular logic gates. The BioBrick database is growing constantly, and the sequences are all freely available.
Features List
- Users must be able to create an account on the website
- Users must be able to search for and find BioBricks in our database
- Users must be able to search by BioBrick ID
- Users must be able to browse BioBricks by category
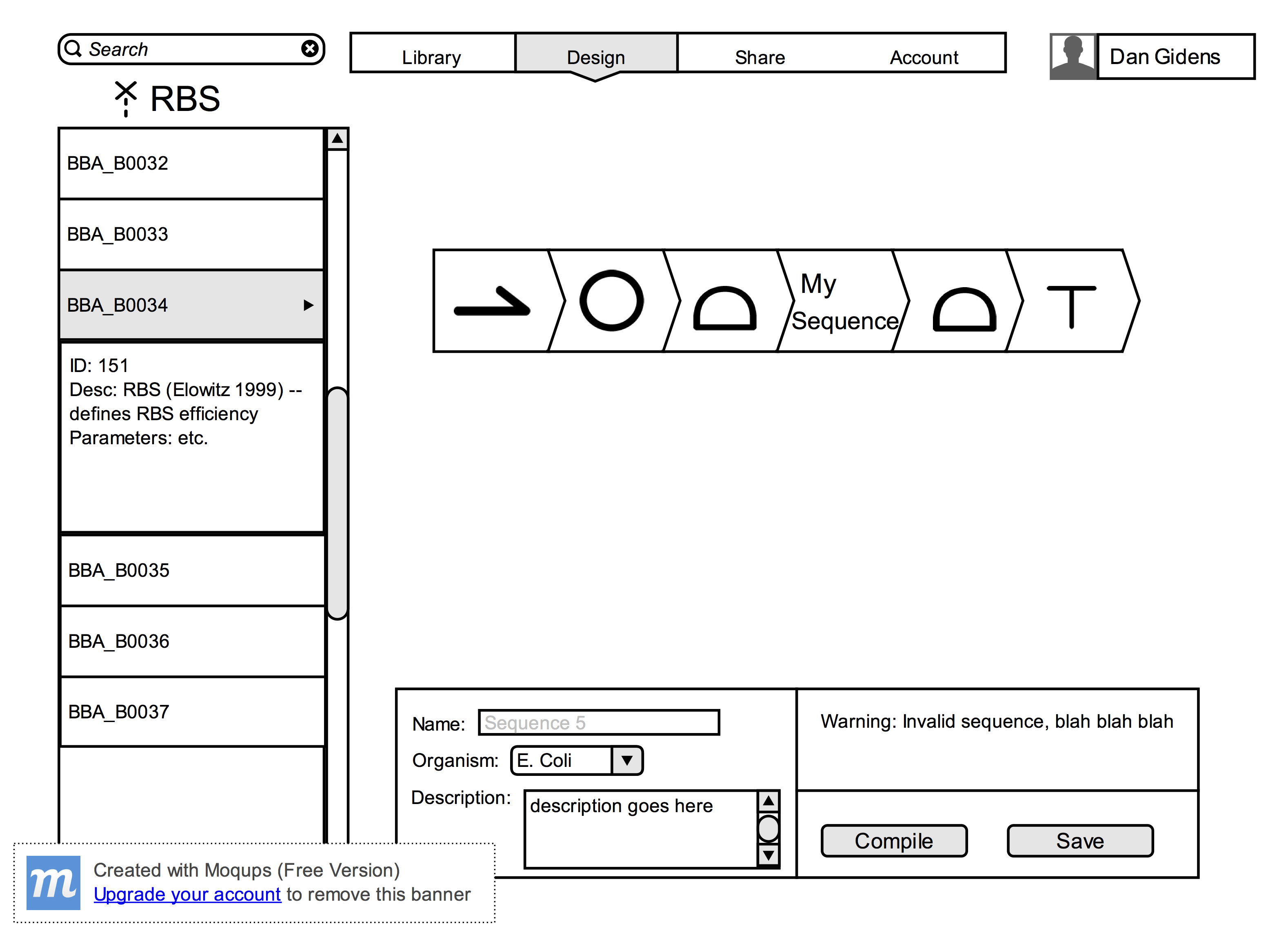
- Users must be able to drag-and-drop BioBricks from the database supply into a design window
- The design window must stitch together the pieces the user has dropped in
- The website must be able to compile the sequence of BioBricks into a DNA sequence
- The website must be able to notify the user of potential problems in the DNA sequence during the compilation
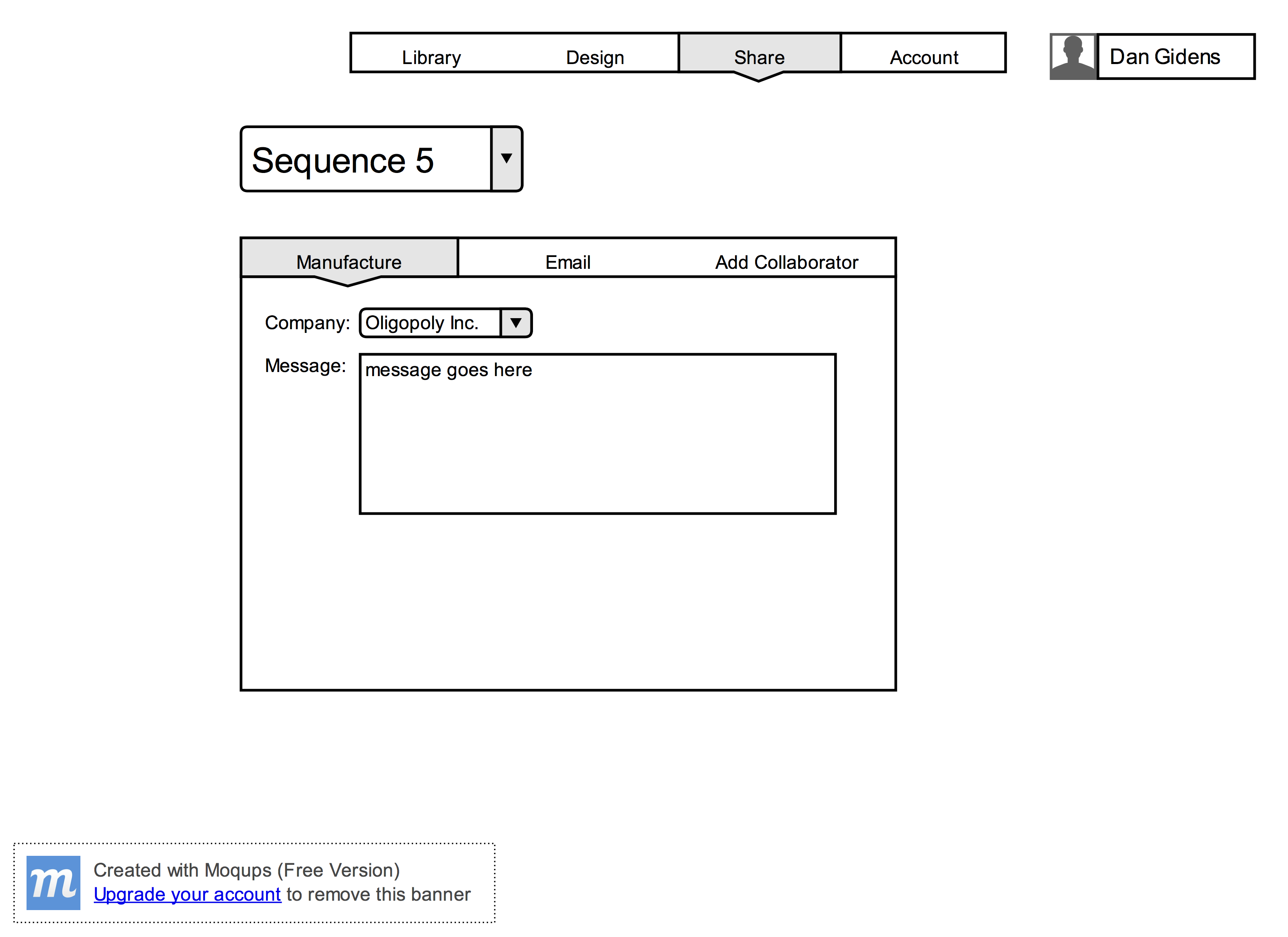
- The user must be able to design specific oligos for their own use
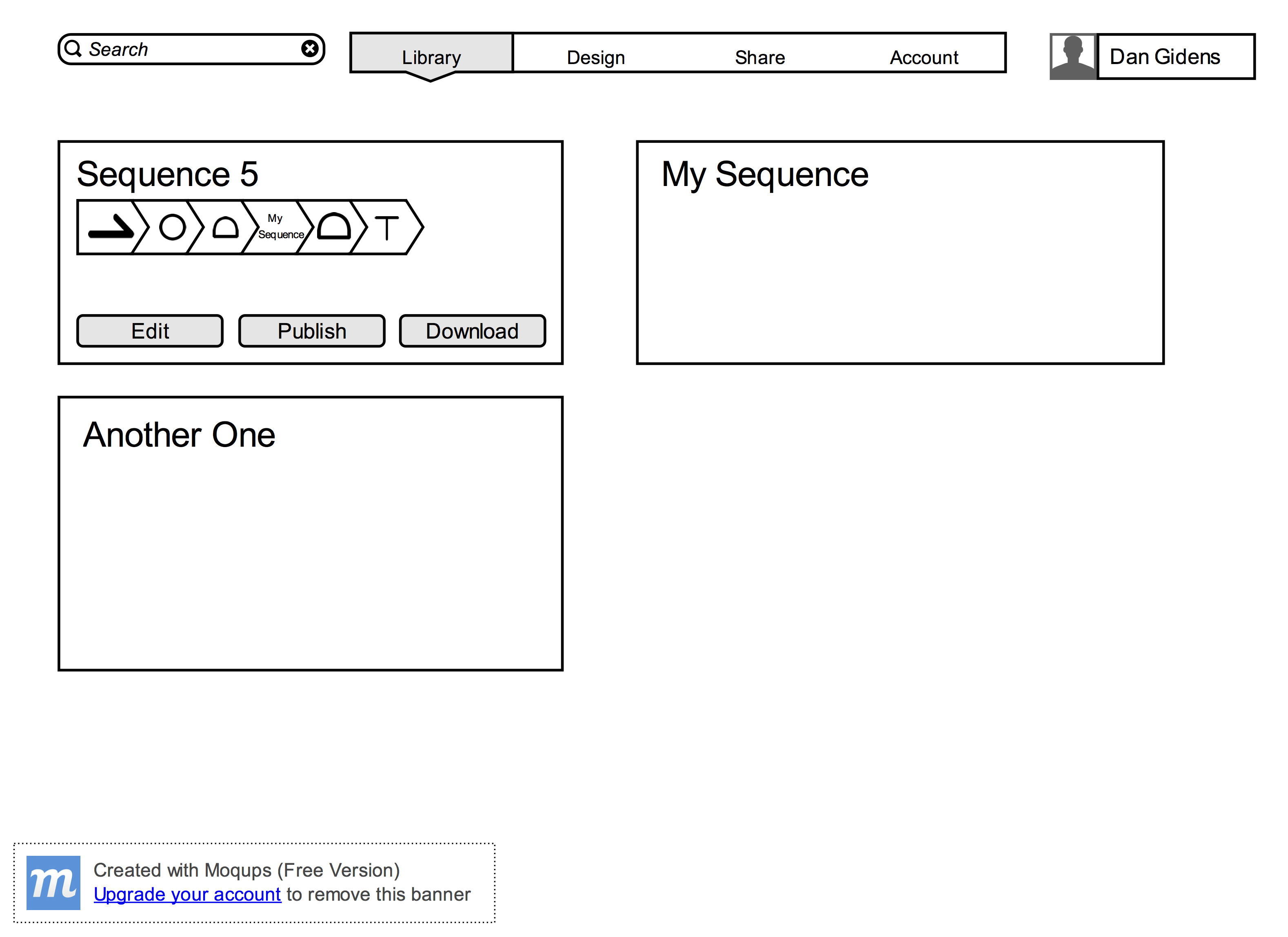
- Users must be able to download the final sequence and/or oligo to their local machines
- Users must be able to email their sequences/oligos to other people
- Users must be able to send out the sequence to an oligo manufacturing company by email
- A User with an account must be able to manage a library of saved custom sequences
- A User with an account must be able to save a sequence to that account’s library
- A User with an account must be able to load a saved sequence into the design window
Extended Features
- Users must be able to share their sequences/oligos with other accounts
- Users must be able to share their sequence with other researchers/collaborators
- The user must be able to specify an organism that the website will codon optimize for when converting the BioBricks to a DNA sequence
Use Cases
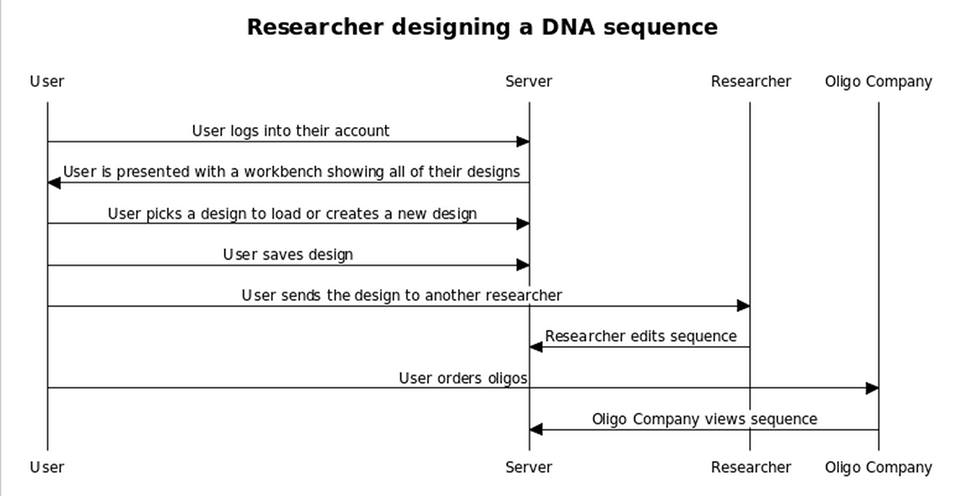
- 1.1 Title: Researcher designing a DNA sequence
- 1.2 Actors: Initiator (the researcher) Server Oligo Company Other researchers
- 1.3 Goal: The researcher wants to be able to design a sequence, save it to their account, design oligos for the sequence, codon optimize the sequence, send the sequence to other researchers for review, and ultimately send the oligo sequences to an oligo company for synthesis
- 1.4 Preconditions: The user has a valid account
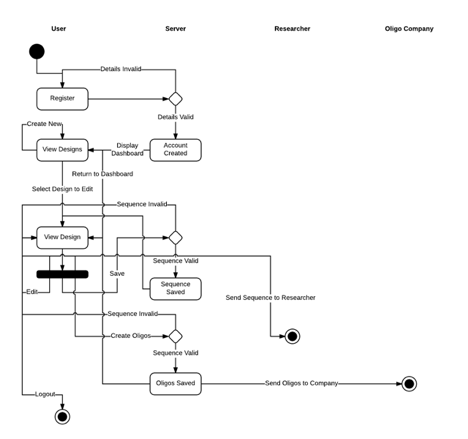
- 1.5 Path: The user logs into their account and is directed to their workbench. Alternate paths at this point: The user selects a previously created design and it is loaded The user decides to create a new design The user then edits the design in front of them, adding, rearranging, and deleting the bioparts in their workspace via a drag and drop interface Alternate paths at this point: The user chooses to save their design and quit The user decides to send a link to their design to a collaborator The user decides to codon optimize their sequence for a particular organism. The sequence will then become associated with an organism, but the association can change later. The user decides to generate oligos for their sequence. This will associate a list of oligos with the sequence, and the oligos will not be changed unless the user decides to re-generate the oligos. Alternate paths at this point: The user may or may not then decide to send the oligos to a sequencing company The user logs out of their account
- 2.1 Title: Researcher views a sequence sent to them by another researcher
- 2.2 Actors: Initiator (a researcher) Server The other researcher
- 2.3 Goal: The researcher wants to view, edit, and/or copy the sequence sent to them by another researcher.
- 2.4 Preconditions: A researcher has sent an email to another researcher indicating that they have created a sequence that needs to be viewed. The second researcher has registered for an account.
- 2.5 Path: The user opens the email sent to them and clicks the link to our site The user logs into their account The user is presented with the sequence. Alternate paths at this point: The user chooses to save the design to their own account and quit The user chooses to edit the design and send a link to the first collaborator. The user decides to send a link to their design to another collaborator The user decides to codon optimize their sequence for a particular organism. The sequence will then become associated with an organism, but the association can change later. The user decides to generate oligos for their sequence. This will associate a list of oligos with the sequence, and the oligos will not be changed unless the user decides to re-generate the oligos. Alternate paths at this point: The user may or may not then decide to send the oligos to a sequencing company The user logs out of their account
- 3.1 Title: Oligo Company views a sequence sent to them
- 3.2 Actors Initiator (a researcher) Oligo Company Server
- 3.3 Goal: The oligo company wants to download the oligo sequences that they are to produce, as well as access information about the user that sent them
- 3.4 Preconditions: A researcher has sent an email to the company indicating that they want to produce the oligos therein
- 3.5 Path: The oligo company opens the email sent to them Alternate paths at this point: The company can simply download the sequences included in the email The company can go to our site and log in There, the company can view the same information online
Javadocs
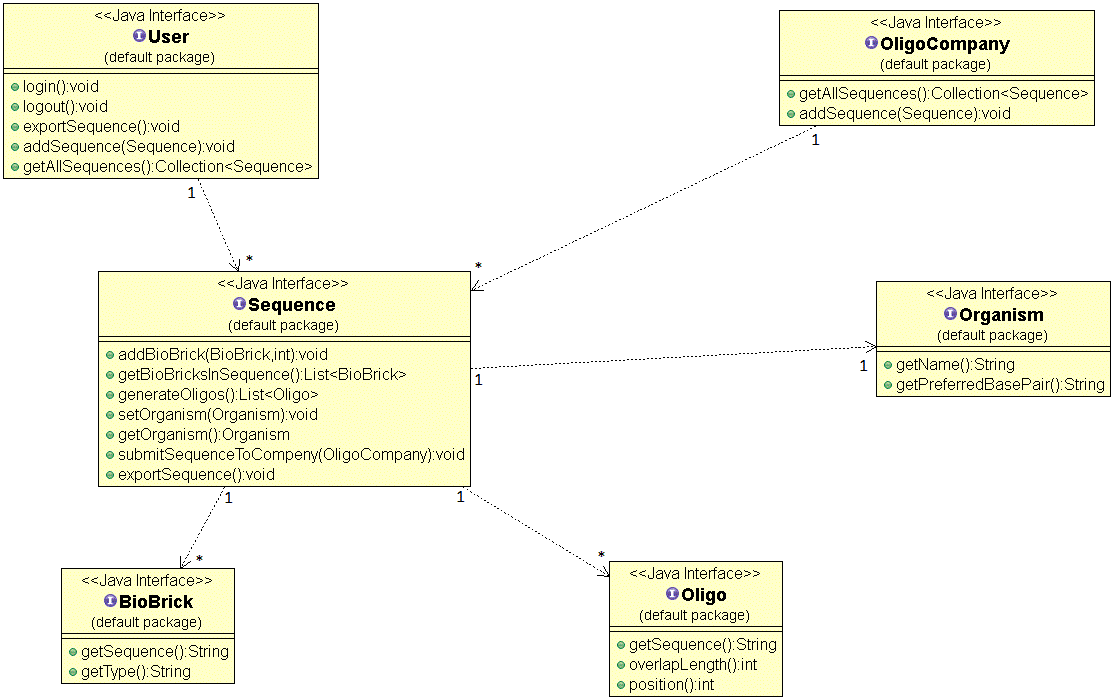
Class Diagram
Courtesy of ObjectAid plugin for Eclipse
Sequence Diagram